This blog will assume you are able to build a package for R and are comfortable with Linux. Fellow STOR-i student Hamish wrote a blog post on how to do this if you haven’t reached this point yet. His post covers how add C++ code to this package also. In this post I will show you how to add documentation for both C++ functions and R functions.
At any level of computer programming you may need help. I have worked with R for quite some time now and mainly use RStudio for its friendly user interface. Despite this experience I still get stuck, forget what a function does, forget what I need to input into a function for it to work, etc.
If this happens in R the quickest way to resolve these issues is to use the “help” function. For example, if I forget what “rnorm” does I can type “help(rnorm)” and it will direct me to the documentation telling me all about what it does. What if this documentation didn’t exist?
I feel that providing documentation for the R packages you make is really important.
Required Packages
To use this tutorial you need both the “Rcpp” package (if you are implementing C++ functions) and “roxygen2” package. In R you can do this using the following code.
install.packages(Rcpp) # Installs the Rcpp package
install.packages(roxygen2) # Installs the roxygen2 packageThe following parts will assume that you have an existing packages skeleton (see Step 1 of Hamish’s blog), with either an R function in the “R” folder of the skeleton or a C++ file in the “src” folder.
First use a text editor of choice to edit the file named “DESCRIPTION”. Simply edit the fields and save. Now navigate into the “man” folder and delete it’s contents, roxygen should generate new documentation files automatically.
Documentation For R Function
To add documentation for an R function open up the folder “R” and find the function you wish to add documentation to. Add the following code above your R function.
#' Name
#'
#' Description
#' @param variable Parameter_Description
#' @return Return_DescriptionReplace “Name” and “Description” with the name of your function and a short description of the function respectively.
“@param” lets roxygen2 know it should add an argument named “variable” (note the naming needs to be the same as in the function). Replace “Parameter_Description” with a short description of the variable. Each variable of the function needs a line with it’s own details.
“@return” lets roxygen2 know it should add a “value” section. Replace “Return_Description” with a short description of what the function outputs.
Documentation For C++ Function
To add documentation for a C++ function open up the folder “src” and find the function you wish to add documentation to. Add the following code above your C++ function (and above the [[Rcpp::export]] line).
//' Name
//'
//' Description
//' @param variable Parameter_Description
//' @return Return_DescriptionAll of these fields are the same as described in the R function section of this tutorial.
Building And Installing The Function
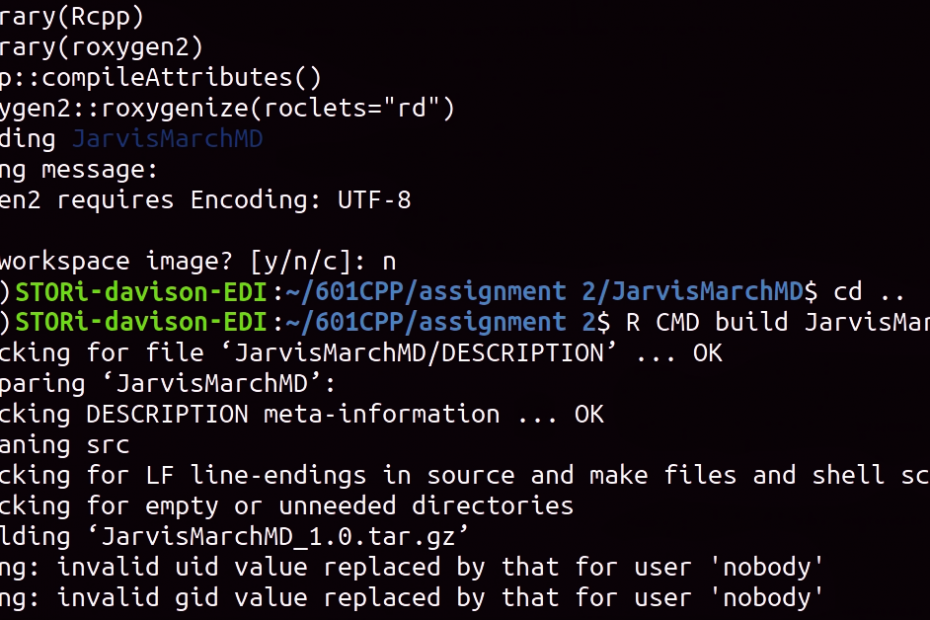
Navigate into the skeleton folder in a Linux terminal window and start R. You need to run the following code.
library(Rcpp)
library(roxygen2)
Rcpp::compileAttributes() # this updates the Rcpp layer from C++ to R
roxygen2::roxygenize(roclets="rd") # this updates the documentationYou can now exit R and move up a folder level using the command “cd ..”
To build the program run the following terminal command making sure to replace “PackageSkeletonName” with the name of your package skeleton. This will produce a package tarball that you can distribute and install.
R CMD build PackageSkeletonNameFor installation follow Step 3 in Hamish’s blog post.
Example Code
The example code contains an algorithm named “Jarvis March” implemented in C++ with documentation that takes a set of 2D points and returns the convex hull, and an R function with documentation that plots the convex hull of a set of 2D points.
The details of these are not important for the tutorial but instead try to spot where the code in this blog has been used.
Example code can be found here
Further Reading
This post written by the authors of Rcpp helped me learn how to add documentation